-Search query
-Search result
Showing 1 - 50 of 5,875 items for (author: ye & j)

EMDB-38328: 
Cryo-EM structure of human GPR1 bound to chemerin
Method: single particle / : Liu AJ, Liu YZ, Ye RD

EMDB-16860: 
Ivabradine bound to HCN4 channel
Method: single particle / : Saponaro A, Chaves-Sanjuan A, Sharifzadeh AS, Clarke OB, Marabelli C, Bolognesi M, Thiel G, Moroni A

PDB-8ofi: 
Ivabradine bound to HCN4 channel
Method: single particle / : Saponaro A, Chaves-Sanjuan A, Sharifzadeh AS, Clarke OB, Marabelli C, Bolognesi M, Thiel G, Moroni A

EMDB-41854: 
Structure of Human Mitochondrial Chaperonin V72I Mutant
Method: single particle / : Chen L, Wang J

PDB-8u39: 
Structure of Human Mitochondrial Chaperonin V72I mutant
Method: single particle / : Chen L, Wang J
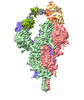
EMDB-37139: 
Structure of SARS-CoV Spike protein complexed with antibody PW5-5
Method: single particle / : Sun L, Mao Q, Wang Y
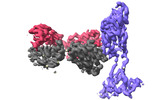
EMDB-37143: 
The local refined map of SARS-CoV-2 XBB Variant Spike protein complexed with antibody PW5-535
Method: single particle / : Sun L, Mao Q, Wang Y
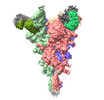
EMDB-37144: 
Trimer state of SARS-CoV Spike protein complexed with antibody PW5-535
Method: single particle / : Sun L, Mao Q, Wang Y

EMDB-37145: 
The local refined map of SARS-CoV Spike protein complexed with antibody PW5-5
Method: single particle / : Sun L, Mao Q, Wang Y
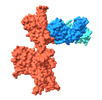
EMDB-37160: 
Monomer state of SARS-CoV-2 XBB Variant Spike protein trimer complexed with antibody PW5-5
Method: single particle / : Sun L, Mao Q, Wang Y
EMDB-37161: 
Monomer state of SARS-CoV Spike protein complexed with antibody PW5-535
Method: single particle / : Sun L, Mao Q, Wang Y
EMDB-37162: 
Structure of SARS-CoV-2 Omicron BA.1 Spike complexed with antibody PW5-570
Method: single particle / : Sun L, Mao Q, Wang Y
EMDB-37163: 
The local refined map of SARS-CoV-2 Omicron BA.1 Spike complexed with antibody PW5-570
Method: single particle / : Sun L, Mao Q, Wang Y
EMDB-37164: 
State 1 of SARS-CoV-2 XBB Variant Spike protein trimer complexed with antibody PW5-5
Method: single particle / : Sun L, Mao Q, Wang Y
EMDB-37165: 
Structure of SARS-CoV-2 XBB Variant Spike protein complexed with broadly neutralizing antibody PW5-535
Method: single particle / : Sun L, Mao Q, Wang Y

PDB-8kdm: 
Structure of SARS-CoV Spike protein complexed with antibody PW5-5
Method: single particle / : Sun L, Mao Q, Wang Y

PDB-8kdr: 
The local refined map of SARS-CoV-2 XBB Variant Spike protein complexed with antibody PW5-535
Method: single particle / : Sun L, Mao Q, Wang Y

PDB-8kds: 
Trimer state of SARS-CoV Spike protein complexed with antibody PW5-535
Method: single particle / : Sun L, Mao Q, Wang Y

PDB-8kdt: 
The local refined map of SARS-CoV Spike protein complexed with antibody PW5-5
Method: single particle / : Sun L, Mao Q, Wang Y

PDB-8kej: 
Monomer state of SARS-CoV-2 XBB Variant Spike protein trimer complexed with antibody PW5-5
Method: single particle / : Sun L, Mao Q, Wang Y

PDB-8kek: 
Monomer state of SARS-CoV Spike protein complexed with antibody PW5-535
Method: single particle / : Sun L, Mao Q, Wang Y

PDB-8keo: 
Structure of SARS-CoV-2 Omicron BA.1 Spike complexed with antibody PW5-570
Method: single particle / : Sun L, Mao Q, Wang Y

PDB-8kep: 
The local refined map of SARS-CoV-2 Omicron BA.1 Spike complexed with antibody PW5-570
Method: single particle / : Sun L, Mao Q, Wang Y

PDB-8keq: 
State 1 of SARS-CoV-2 XBB Variant Spike protein trimer complexed with antibody PW5-5
Method: single particle / : Sun L, Mao Q, Wang Y

PDB-8ker: 
Structure of SARS-CoV-2 XBB Variant Spike protein complexed with broadly neutralizing antibody PW5-535
Method: single particle / : Sun L, Mao Q, Wang Y

EMDB-41849: 
Structure of 310-18A5 Fab in complex with A/Solomon Islands/3/2006(H1N1) influenza virus hemagglutinin
Method: single particle / : Lei R, Wu NC

EMDB-43835: 
Structure of biofilm-forming functional amyloid PSMa1 from Staphylococcus aureus
Method: helical / : Hansen KH, Byeon CH, Liu Q, Drace T, Boesen T, Conway JF, Andreasen M, Akbey U

EMDB-50222: 
Human DNA polymerase epsilon bound to DNA and PCNA (open conformation)
Method: single particle / : Roske JJ, Yeeles JTP

EMDB-50223: 
Human DNA polymerase epsilon bound to DNA and PCNA (ajar conformation)
Method: single particle / : Roske JJ, Yeeles JTP

EMDB-50224: 
Human DNA polymerase epsilon bound to DNA and PCNA (closed conformation)
Method: single particle / : Roske JJ, Yeeles JTP

EMDB-50225: 
Human DNA Polymerase epsilon bound to T-C mismatched DNA (Post-Insertion state)
Method: single particle / : Roske JJ, Yeeles JTP

EMDB-50226: 
Human DNA Polymerase epsilon bound to T-C mismatched DNA (Polymerase Arrest state)
Method: single particle / : Roske JJ, Yeeles JTP

EMDB-50227: 
Human DNA Polymerase epsilon bound to T-C mismatched DNA (Frayed Substrate state)
Method: single particle / : Roske JJ, Yeeles JTP

EMDB-50228: 
Human DNA Polymerase epsilon bound to T-C mismatched DNA (Mismatch Excision state)
Method: single particle / : Roske JJ, Yeeles JTP

EMDB-60655: 
Orf virus scaffolding protein Orfv075
Method: single particle / : Hyun J, Kim S, Ko S, Kim M, Jang Y

PDB-9ikc: 
Orf virus scaffolding protein Orfv075
Method: single particle / : Hyun J, Kim S, Ko S, Kim M, Jang Y

PDB-9atw: 
Structure of biofilm-forming functional amyloid PSMa1 from Staphylococcus aureus
Method: helical / : Hansen KH, Byeon CH, Liu Q, Drace T, Boesen T, Conway JF, Andreasen M, Akbey U

PDB-9f6d: 
Human DNA polymerase epsilon bound to DNA and PCNA (open conformation)
Method: single particle / : Roske JJ, Yeeles JTP

PDB-9f6e: 
Human DNA polymerase epsilon bound to DNA and PCNA (ajar conformation)
Method: single particle / : Roske JJ, Yeeles JTP

PDB-9f6f: 
Human DNA polymerase epsilon bound to DNA and PCNA (closed conformation)
Method: single particle / : Roske JJ, Yeeles JTP

PDB-9f6i: 
Human DNA Polymerase epsilon bound to T-C mismatched DNA (Post-Insertion state)
Method: single particle / : Roske JJ, Yeeles JTP

PDB-9f6j: 
Human DNA Polymerase epsilon bound to T-C mismatched DNA (Polymerase Arrest state)
Method: single particle / : Roske JJ, Yeeles JTP

PDB-9f6k: 
Human DNA Polymerase epsilon bound to T-C mismatched DNA (Frayed Substrate state)
Method: single particle / : Roske JJ, Yeeles JTP

PDB-9f6l: 
Human DNA Polymerase epsilon bound to T-C mismatched DNA (Mismatch Excision state)
Method: single particle / : Roske JJ, Yeeles JTP

EMDB-36914: 
Cryo-EM structure of Streptomyces coelicolor transcription initiation complex with the global transcription factor AfsR
Method: single particle / : Lin W, Shi J, Xu JC

EMDB-41578: 
mGluR3 class 1 in the presence of the antagonist LY 341495
Method: single particle / : Strauss A, Levitz J

EMDB-45242: 
mGluR3 in the presence of the antagonist LY 341495 and positive allosteric modulator VU6023326 class 2
Method: single particle / : Strauss A, Levitz J

PDB-8trd: 
mGluR3 class 1 in the presence of the antagonist LY 341495
Method: single particle / : Strauss A, Levitz J

EMDB-18538: 
p97 in DNA origami cage
Method: single particle / : Manar E, Amelie HJ

EMDB-37515: 
Cryo-EM structure of inward-open state human norepinephrine transporter NET bound with antidepressant desipramine in KCl condition.
Method: single particle / : Tan J, Xiao Y, Kong F, Lei J, Yuan Y, Yan C
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
